dcmri.TissueArray#
- class dcmri.TissueArray(shape, kinetics='HF', water_exchange='FF', sequence='SS', aif=None, ca=None, t=None, dt=1.0, free=None, parallel=False, verbose=0, **params)[source]#
Pixel-based vascular-interstitial tissue.
This is the most common tissue type as found in for instance brain, cancer, lung, muscle, prostate, skin, and more. For more detail see Exchange tissues.
Usage of this class is mostly identical to
dcmri.Tissue.- Parameters:
shape (array-like) – shape of the tissue array in dimensions. Any number of dimensions is allowed but the last must be time.
kinetics (str, optional) – Tracer-kinetic model. Possible values are ‘2CX’, ‘2CU’, ‘HF’, ‘HFU’, ‘NX’, ‘FX’, ‘WV’, ‘U’. Defaults to ‘HF’.
water_exchange (str, optional) – Water exchange regime, Any combination of two of the letters ‘F’, ‘N’, ‘R’ is allowed. Defaults to ‘FF’.
sequence (str, optional) – imaging sequence. Possible values are ‘SS’ and ‘SR’. Defaults to ‘SS’.
aif (array-like, optional) – Signal-time curve in the blood of the feeding artery. If aif is not provided, the arterial blood concentration is ca. Defaults to None.
ca (array-like, optional) – Blood concentration in the arterial input. ca is ignored if aif is provided, but is required otherwise. Defaults to None.
t (array-like, optional) – Time points of the arterial input function. If t is not provided, the temporal sampling is uniform with interval dt. Defaults to None.
dt (float, optional) – Time interval between values of the arterial input function. dt is ignored if t is provided. Defaults to 1.0.
free (dict, optional) – Dictionary with free parameters and their bounds. If not provided, a default set of free parameters is used. Defaults to None.
parallel (bool, optional) – If True, computations are parallelized. Defaults to False.
verbose (int, optional) – verbosity of the computation. With verbose=0, no feedback is given; with verbose=1, a status bar is shown. Defaults to 0.
params (dict, optional) – values for the parameters of the tissue, specified as keyword parameters. Defaults are used for any that are not provided. The parameters are the same as in
dcmri.Tissue.
See also
Example
Fit a coarse image of the brain using a 2-compartment exchange model.
>>> import numpy as np >>> import dcmri as dc
Use
fake_brainto generate synthetic test data:>>> n=8 >>> time, signal, aif, gt = dc.fake_brain(n)
Build a tissue array and set the constants to match the experimental conditions of the synthetic test data:
>>> shape = (n,n) >>> tissue = dc.TissueArray( ... shape, ... kinetics = '2CX', ... aif = aif, ... dt = time[1], ... r1 = dc.relaxivity(3, 'blood', 'gadodiamide'), ... TR = 0.005, ... FA = 15, ... R10a = 1/dc.T1(3.0,'blood'), ... R10 = 1/gt['T1'], ... n0 = 10, ... )
Train the tissue on the data:
>>> tissue.train(time, signal)
Plot the reconstructed maps, along with their standard deviations and the ground truth for reference:
>>> tissue.plot(time, signal, ref=gt)
(
Source code,png,hires.png,pdf)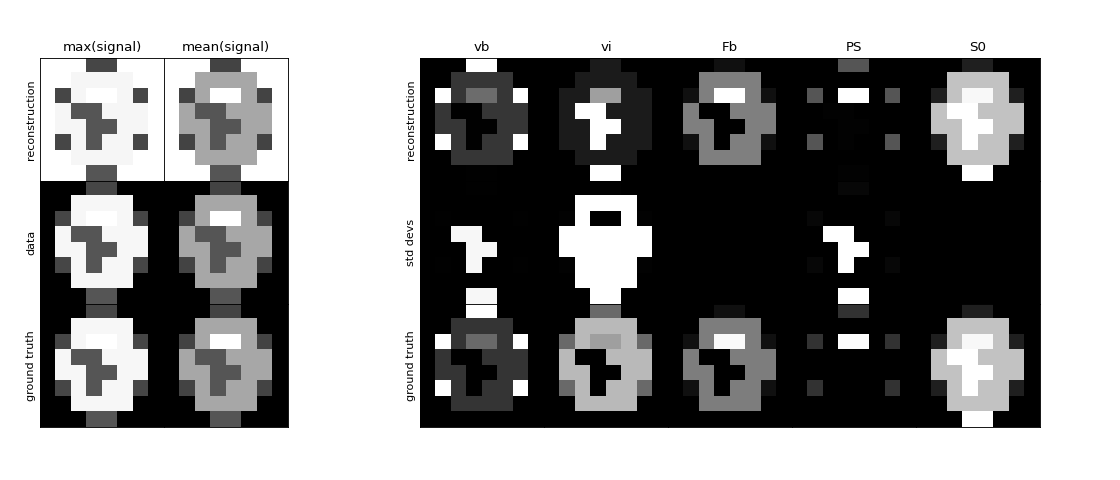
Methods
cost(time, signal[, metric])Return the goodness-of-fit
Model parameters with descriptions.
info()Print detailed information about the tissue
load([file, path, filename])Load the saved state of the model
params(*args[, round_to])Return the parameter values
plot(time, signal[, vmin, vmax, cmap, ref, ...])Plot parameter maps
plot_fit(time, signal[, hist_kwargs, roi, ...])Plot time curves and fits in representative pixels.
plot_params([roi, vmin, vmax, ref, fname, show])Show parameter distributions in regions of interest.
plot_signals(time, signal[, cmap, ref, ...])Plot measured and reconstructed dynamic signals.
predict(time)Predict the data at given time points
save([file, path, filename])Save the current state of the model
set_free([pop])Set the free model parameters.
train(time, signal, **kwargs)Train the free parameters
- cost(time, signal, metric='NRMS') float[source]#
Return the goodness-of-fit
- Parameters:
time (array-like) – Array with time points.
signal (array-like) – Array with measured signals for each element of time.
metric (str, optional) – Which metric to use. Possible metrics are ‘RMS’ (Root-mean-square); ‘NRMS’ (Normalized root-mean-square); ‘AIC’ (Akaike information criterion); ‘cAIC’ (Corrected Akaike information criterion for small models); ‘BIC’ (Bayesian information criterion). Defaults to ‘NRMS’.
- Returns:
goodness of fit in each element of the data array.
- Return type:
ndarray
- export_params()[source]#
Model parameters with descriptions.
- Returns:
- Dictionary with one item for each model parameter. The key
is the parameter symbol (short name), and the value is a 4-element list with [parameter name, value, unit, sdev].
- Return type:
Example
List the parameters that are exported for a given model:
>>> import dcmri as dc >>> tissue = dc.TissueArray((8,8), ... kinetics = '2CU', ... water_exchange = 'FR', ... ) >>> print(tissue.export_params().keys()) dict_keys(['PSc', 'H', 'vb', 'vi', 'Fb', 'PS', 'Fp', 'vp', 'Ktrans', 'E', 'Ti', 'Tp', 'Tb', 'Twc', 'FAcorr'])
- info()[source]#
Print detailed information about the tissue
Example
List all parameters of a default tissue:
>>> import dcmri as dc >>> tissue = dc.Tissue() >>> tissue.info() ------------- Configuration ------------- Kinetics: HF Water exchange regime: FF Imaging sequence: SS ---------- Parameters ---------- r1 --> Full name: Contrast agent relaxivity --> Units: Hz/M --> Initial value: 5000.0 --> Current value: 5000.0 --> Free parameter: No --> Bounds: [0, inf] R10a --> Full name: Arterial precontrast R1 --> Units: Hz --> Initial value: 0.7 --> Current value: 0.7 --> Free parameter: No --> Bounds: [0, inf] B1corr_a --> Full name: Arterial B1-correction factor --> Units: --> Initial value: 1 --> Current value: 1 --> Free parameter: No --> Bounds: [0, inf] S0 --> Full name: Signal scaling factor --> Units: a.u. --> Initial value: 1.0 --> Current value: 1.0 --> Free parameter: No --> Bounds: [0, inf] B1corr --> Full name: Tissue B1-correction factor --> Units: --> Initial value: 1 --> Current value: 1 --> Free parameter: No --> Bounds: [0, inf] FA --> Full name: Flip angle --> Units: deg --> Initial value: 15 --> Current value: 15 --> Free parameter: No --> Bounds: [0, inf] TR --> Full name: Repetition time --> Units: sec --> Initial value: 0.005 --> Current value: 0.005 --> Free parameter: No --> Bounds: [0, inf] TS --> Full name: Sampling time --> Units: sec --> Initial value: 0 --> Current value: 0 --> Free parameter: No --> Bounds: [0, inf] H --> Full name: Tissue Hematocrit --> Units: --> Initial value: 0.45 --> Current value: 0.45 --> Free parameter: No --> Bounds: [0.001, 0.999] vb --> Full name: Blood volume --> Units: mL/cm3 --> Initial value: 0.1 --> Current value: 0.1 --> Free parameter: Yes --> Bounds: [0.001, 0.999] vi --> Full name: Interstitial volume --> Units: mL/cm3 --> Initial value: 0.3 --> Current value: 0.3 --> Free parameter: Yes --> Bounds: [0.001, 0.999] PS --> Full name: Permeability-surface area product --> Units: mL/sec/cm3 --> Initial value: 0.003 --> Current value: 0.003 --> Free parameter: Yes --> Bounds: [0, inf] R10 --> Full name: Tissue precontrast R1 --> Units: Hz --> Initial value: 0.7 --> Current value: 0.7 --> Free parameter: No --> Bounds: [0, inf] n0 --> Full name: Number of precontrast acquisitions --> Units: --> Initial value: 1 --> Current value: 1 --> Free parameter: No
- load(file=None, path=None, filename='Model')#
Load the saved state of the model
- Parameters:
file (str, optional) – complete path of the file. If this is not provided, a file is constructure from path amd filename variables. Defaults to None.
path (str, optional) – path to store the state if file is not provided. This variable is ignored if file is provided. Defaults to current working directory.
filename (str, optional) – filename to store the state if file is not provided. If no extension is included, the extension ‘.pkl’ is automatically added. This variable is ignored if file is provided. Defaults to ‘Model’.
- Returns:
class instance
- Return type:
- plot(time, signal, vmin={}, vmax={}, cmap='gray', ref=None, fname=None, show=True)[source]#
Plot parameter maps
- Parameters:
time (array-like) – 1D array with time points.
signal (array-like) – Array of signal curves. Any number of dimensions is allowed but the last dimension is time.
vmin (dict, optional) – Minimum values on display for given parameters. Defaults to {}.
vmax (dict, optional) – Maximum values on display for given parameters. Defaults to {}.
cmap (str, optional) – matplotlib colormap. Defaults to ‘gray’.
ref (dict, optional) – Reference images - typically used to display ground truth data when available. Keys are ‘signal’ (array of data in the same shape as signal), and the parameter maps to show. Defaults to None.
fname (str, optional) – File path to save image. Defaults to None.
show (bool, optional) – Determine whether the image is shown or not. Defaults to True.
- Raises:
NotImplementedError – Features that are not currently implemented.
- plot_fit(time, signal, hist_kwargs={}, roi=None, ref=None, fname=None, show=True)[source]#
Plot time curves and fits in representative pixels.
- Parameters:
time (array-like) – 1D array with time points.
signal (array-like) – Array of signal curves. Any number of dimensions is allowed but the last dimension is time.
hist_kwargs (dict, optional) – Keyword arguments to be passed on the matlotlib’s hist() finction. Defaults to {}.
roi (dict, optional) – Dictionary with masks for regions-of-interest to be shown in the plot. if none is provided, the entire array is shown. Defaults to None.
ref (dict, optional) – Reference images - typically used to display ground truth data when available. Keys are ‘signal’ (array of data in the same shape as signal), and the parameter maps to show. Defaults to None.
fname (str, optional) – File path to save image. Defaults to None.
show (bool, optional) – Determine whether the image is shown or not. Defaults to True.
- plot_params(roi=None, vmin={}, vmax={}, ref=None, fname=None, show=True)[source]#
Show parameter distributions in regions of interest.
- Parameters:
roi (dict, optional) – Dictionary with masks for regions-of-interest to be shown in the plot. if none is provided, the entire array is shown. Defaults to None.
vmin (dict, optional) – Minimum values on display for given parameters. Defaults to {}.
vmax (dict, optional) – Maximum values on display for given parameters. Defaults to {}.
cmap (str, optional) – matplotlib colormap. Defaults to ‘gray’.
ref (dict, optional) – Reference images - typically used to display ground truth data when available. Keys are ‘signal’ (array of data in the same shape as ydata), and the parameter maps to show. Defaults to None.
fname (str, optional) – File path to save image. Defaults to None.
show (bool, optional) – Determine whether the image is shown or not. Defaults to True.
- plot_signals(time, signal, cmap='gray', ref=None, fname=None, show=True)[source]#
Plot measured and reconstructed dynamic signals.
- Parameters:
time (array-like) – 1D array with time points.
signal (array-like) – Array of signal curves. Any number of dimensions is allowed but the last dimension is time.
cmap (str, optional) – matplotlib colormap. Defaults to ‘gray’.
ref (dict, optional) – Reference images - typically used to display ground truth data when available. Keys are ‘signal’ (array of data in the same shape as signal), and the parameter maps to show. Defaults to None.
fname (str, optional) – File path to save image. Defaults to None.
show (bool, optional) – Determine whether the image is shown or not. Defaults to True.
- Raises:
NotImplementedError – Features that are not currently implemented.
- predict(time: ndarray) ndarray[source]#
Predict the data at given time points
- Parameters:
time (array-like) – 1D array with time points.
- Returns:
Array of predicted y-values.
- Return type:
ndarray
- save(file=None, path=None, filename='Model')#
Save the current state of the model
- Parameters:
file (str, optional) – complete path of the file. If this is not provided, a file is constructure from path amd filename variables. Defaults to None.
path (str, optional) – path to store the state if file is not provided. This variable is ignored if file is provided. Defaults to current working directory.
filename (str, optional) – filename to store the state if file is not provided. If no extension is included, the extension ‘.pkl’ is automatically added. This variable is ignored if file is provided. Defaults to ‘Model’.
- Returns:
class instance
- Return type:
- set_free(pop=None, **kwargs)#
Set the free model parameters.
- Parameters:
pop (str or list) – a single variable or a list of variables to remove from the list of free parameters.
- Raises:
ValueError – if the pop argument contains a parameter that is not in the list of free parameters.
ValueError – If the parameter is not a model parameter, or bounds are not properly formatted.
- train(time: ndarray, signal: ndarray, **kwargs)[source]#
Train the free parameters
- Parameters:
time (array-like) – 1D array with time points.
signal (array-like) – Array of signal curves. Any number of dimensions is allowed but the last dimension must be time.
kwargs – any keyword parameters accepted by
Tissue.train.
- Returns:
A reference to the model instance.
- Return type:
