dcmri.Aorta#
- class dcmri.Aorta(organs='2cxm', heartlung='pfcomp', sequence='SS', **params)[source]#
Whole-body model for aorta signal.
The model represents the body as a leaky loop with a heart-lung system and an organ system. The heart-lung system is modelled as a chain compartment and the organs are modelled as a two-compartment exchange model. Bolus injection into the system is modelled as a step function.
Injection parameters
weight (float, default=70): Subject weight in kg.
agent (str, default=’gadoterate’): Contrast agent generic name.
dose (float, default=0.2): Injected contrast agent dose in mL per kg bodyweight.
rate (float, default=1): Contrast agent injection rate in mL per sec.
Acquisition parameters
sequence (str, default=’SS’): Signal model.
tmax (float, default=120): Maximum acquisition time in sec.
field_strength (float, default=3.0): Magnetic field strength in T.
t0 (float, default=1): Baseline length in secs.
TR (float, default=0.005): Repetition time, or time between excitation pulses, in sec.
FA (float, default=15): Nominal flip angle in degrees.
TC (float, default=0.1): Time to the center of k-space in a saturation-recovery sequence (sec).
Signal parameters
R10 (float, default=1): Precontrast arterial relaxation rate in 1/sec.
S0 (float, default=1): scale factor for the arterial MR signal in the first scan.
Whole body kinetic parameters
heartlung (str, default=’pfcomp’): Kinetic model for the heart-lung system (either ‘pfcomp’ or ‘chain’).
organs (str, default=’2cxm’): Kinetic model for the organs.
BAT (float, default=60): Bolus arrival time, i.e. time point where the indicator first arrives in the body.
BAT2 (float, default=1200): Bolus arrival time in the second scan, i.e. time point where the indicator first arrives in the body.
CO (float, default=100): Cardiac output in mL/sec.
Thl (float, default=10): Mean transit time through heart and lungs.
Dhl (float, default=0.2): Dispersion through the heart-lung system, with a value in the range [0,1].
To (float, default=20): average time to travel through the organ’s vasculature.
Eb (float, default=0.05): fraction of indicator extracted from the vasculature in a single pass.
Eo (float, default=0.15): Fraction of indicator entering the organs which is extracted from the blood pool.
Teb (float, default=120): Average time to travel through the organs extravascular space.
Prediction and training parameters
dt (float, default=1): Internal time resolution of the AIF in sec.
dose_tolerance (fload, default=0.1): Stopping criterion for the whole-body model.
free (array-like): list of free parameters. The default depends on the kinetics parameter.
free (array-like): 2-element list with lower and upper free of the free parameters. The default depends on the kinetics parameter.
- Parameters:
params (dict, optional) – override defaults for any of the parameters.
See also
Example
Use the model to reconstruct concentrations from experimentally derived signals.
>>> import matplotlib.pyplot as plt >>> import dcmri as dc
Use
fake_tissueto generate synthetic test data from experimentally-derived concentrations:>>> time, aif, _, gt = dc.fake_tissue()
Build an aorta model and set weight, contrast agent, dose and rate to match the conditions of the original experiment (Parker et al 2006):
>>> aorta = dc.Aorta( ... dt = 1.5, ... weight = 70, ... agent = 'gadodiamide', ... dose = 0.2, ... rate = 3, ... field_strength = 3.0, ... TR = 0.005, ... FA = 15, ... R10 = 1/dc.T1(3.0,'blood'), ... )
Train the model on the data:
>>> aorta.train(time, aif)
Plot the reconstructed signals and concentrations and compare against the experimentally derived data:
>>> aorta.plot(time, aif)
We can also have a look at the model parameters after training:
>>> aorta.print_params(round_to=3) ----------------------------------------- Free parameters with their errors (stdev) ----------------------------------------- Bolus arrival time (BAT): 18.485 (5.656) sec Cardiac output (CO): 228.237 (29.321) mL/sec Heart-lung mean transit time (Thl): 9.295 (6.779) sec Heart-lung transit time dispersion (Dhl): 0.459 (0.177) Organs mean transit time (To): 29.225 (11.646) sec Extraction fraction (Eb): 0.013 (0.972) Organs extraction fraction (Eo): 0.229 (0.582) Extracellular mean transit time (Te): 97.626 (640.454) sec ------------------ Derived parameters ------------------ Mean circulation time (Tc): 38.521 sec
Note: The extracellular mean transit time has a high error, indicating that the acquisition time here is insufficient to resolve the transit through the leakage space.
(
Source code,png,hires.png,pdf)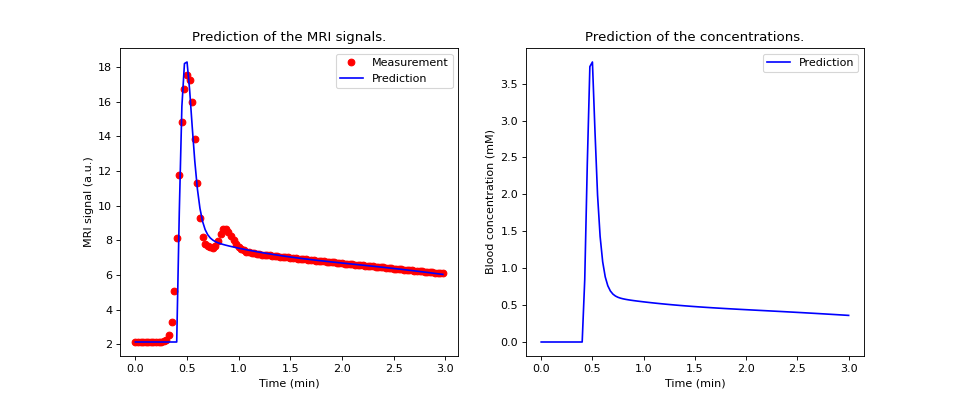
Attributes
lower- and upper free for all free parameters.
Methods
conc()Aorta blood concentration
cost(xdata, ydata[, metric])Return the goodness-of-fit
Return model parameters with their descriptions
load([file, path, filename])Load the saved state of the model
params(*args[, round_to])Return the parameter values
plot(xdata, ydata[, ref, xlim, fname, show])Plot the model fit against data
predict(xdata)Predict the data at given xdata
print_params([round_to])Print the model parameters and their uncertainties
relax()Aorta longitudinal relation rate
save([file, path, filename])Save the current state of the model
set_free([pop])Set the free model parameters.
train(xdata, ydata, **kwargs)Train the free parameters
- conc()[source]#
Aorta blood concentration
- Parameters:
t (array-like) – Time points of the concentration (sec)
- Returns:
Concentration in M
- Return type:
- cost(xdata, ydata, metric='NRMS') float#
Return the goodness-of-fit
- Parameters:
xdata (array-like) – Array with x-data (time points).
ydata (array-like) – Array with y-data (signal values)
metric (str, optional) – Which metric to use (see notes for possible values). Defaults to ‘NRMS’.
- Returns:
goodness of fit.
- Return type:
Notes
Available options are:
‘RMS’: Root-mean-square.
‘NRMS’: Normalized root-mean-square.
‘AIC’: Akaike information criterion.
‘cAIC’: Corrected Akaike information criterion for small models.
‘BIC’: Baysian information criterion.
- export_params()[source]#
Return model parameters with their descriptions
- Returns:
Dictionary with one item for each model parameter. The key is the parameter symbol (short name), and the value is a 4-element list with [parameter name, value, unit, sdev].
- Return type:
- free = {}#
lower- and upper free for all free parameters.
- load(file=None, path=None, filename='Model')#
Load the saved state of the model
- Parameters:
file (str, optional) – complete path of the file. If this is not provided, a file is constructure from path and filename variables. Defaults to None.
path (str, optional) – path to store the state if file is not provided. Thos variable is ignored if file is provided. Defaults to current working directory.
filename (str, optional) – filename to store the state if file is not provided. If no extension is included, the extension ‘.pkl’ is automatically added. This variable is ignored if file is provided. Defaults to ‘Model’.
- Returns:
class instance
- Return type:
- params(*args, round_to=None)#
Return the parameter values
- plot(xdata: ndarray, ydata: ndarray, ref=None, xlim=None, fname=None, show=True)[source]#
Plot the model fit against data
- Parameters:
xdata (array-like) – Array with x-data (time points)
ydata (array-like) – Array with y-data (signal data)
xlim (array_like, optional) – 2-element array with lower and upper boundaries of the x-axis. Defaults to None.
ref (tuple, optional) – Tuple of optional test data in the form (x,y), where x is an array with x-values and y is an array with y-values. Defaults to None.
fname (path, optional) – Filepath to save the image. If no value is provided, the image is not saved. Defaults to None.
show (bool, optional) – If True, the plot is shown. Defaults to True.
- predict(xdata) ndarray[source]#
Predict the data at given xdata
- Parameters:
xdata (array-like) – Either an array with x-values (time points) or a tuple with multiple such arrays
- Returns:
- Either an array of predicted y-values (if
xdata is an array) or a tuple of such arrays (if xdata is a tuple).
- Return type:
tuple or array-like
- print_params(round_to=None)#
Print the model parameters and their uncertainties
- Parameters:
round_to (int, optional) – Round to how many digits. If this is not provided, the values are not rounded. Defaults to None.
- relax()[source]#
Aorta longitudinal relation rate
- Parameters:
t (array-like) – Time points of the concentration (sec)
- Returns:
Concentration in M
- Return type:
- save(file=None, path=None, filename='Model')#
Save the current state of the model
- Parameters:
file (str, optional) – complete path of the file. If this is not provided, a file is constructure from path and filename variables. Defaults to None.
path (str, optional) – path to store the state if file is not provided. Thos variable is ignored if file is provided. Defaults to current working directory.
filename (str, optional) – filename to store the state if file is not provided. If no extension is included, the extension ‘.pkl’ is automatically added. This variable is ignored if file is provided. Defaults to ‘Model’.
- Returns:
class instance
- Return type:
- set_free(pop=None, **kwargs)#
Set the free model parameters.
- Parameters:
pop (str or list) – a single variable or a list of variables to remove from the list of free parameters.
- Raises:
ValueError – if the pop argument contains a parameter that is not in the list of free parameters.
ValueError – If the parameter is not a model parameter, or bounds are not properly formatted.
- train(xdata, ydata, **kwargs)[source]#
Train the free parameters
- Parameters:
xdata (array-like) – Array with x-data (time points)
ydata (array-like) – Array with y-data (signal data)
kwargs – any keyword parameters accepted by
scipy.optimize.curve_fit.
- Returns:
A reference to the model instance.
- Return type:
Model
