dcmri.Liver#
- class dcmri.Liver(kinetics='2I-EC', stationary='UE', sequence='SS', config=None, aif=None, ca=None, vif=None, cv=None, t=None, dt=0.5, free=None, **params)[source]#
General model for liver tissue.
This is the standard interface for liver tissues with known input function(s). For more detail see Liver.
- Parameters:
kinetics (str, optional) – Tracer-kinetic model. See table Kinetic models for the liver for options. Defaults to ‘2I-EC’.
stationary (str, optional) – For intracellular tracers - stationarity regime of the hepatocytes. The options are ‘UE’, ‘E’, ‘U’ or None. For more detail see Liver. Defaults to ‘UE’.
sequence (str, optional) – imaging sequence. Possible values are ‘SS’ and ‘SR’. Defaults to ‘SS’.
config (str, optional) – configuration option for using pre-defined variable values from use cases. Currently, the available options for this are ‘TRISTAN-rat’. Defaults to None.
aif (array-like, optional) – Signal-time curve in the blood of the feeding artery. If aif is not provided, the arterial blood concentration is ca. Defaults to None.
ca (array-like, optional) – Blood concentration in the arterial input. ca is ignored if aif is provided, but is required otherwise. Defaults to None.
vif (array-like, optional) – Signal-time curve in the blood of the portal vein. If vif is not provided, the venous blood concentration is cv. Defaults to None.
cv (array-like, optional) – Blood concentration in the portal venous input. cv is ignored if vif is provided, but is required otherwise. Defaults to None.
t (array-like, optional) – Time points of the arterial input function. If t is not provided, the temporal sampling is uniform with interval dt. Defaults to None.
dt (float, optional) – Time interval between values of the arterial input function. dt is ignored if t is provided. Defaults to 1.0.
free (dict, optional) – Dictionary with free parameters and their bounds. If not provided, a default set of free parameters is used. Defaults to None.
params (dict, optional) – values for the parameters of the tissue, specified as keyword parameters. Defaults are used for any that are not provided. See tables Liver parameters and Liver parameter defaults for a list of parameters and their default values.
See also
Example
Fit a dual-inlet liver model:
>>> import matplotlib.pyplot as plt >>> import dcmri as dc
Use
fake_liverto generate synthetic test data:>>> time, aif, vif, roi, gt = dc.fake_liver()
Build a tissue model and set the constants to match the experimental conditions of the synthetic test data. Note the default model is the dual-inlet model for extracellular agents (2I-EC). Since the synthetic data are generated with an intracellular agent, the default for the kinetic model needs to be overwritten:
>>> model = dc.Liver( ... kinetics = '2I-IC', ... aif = aif, ... vif = vif, ... dt = time[1], ... agent = 'gadoxetate', ... field_strength = 3.0, ... TR = 0.005, ... FA = 15, ... n0 = 10, ... R10 = 1/dc.T1(3.0,'liver'), ... R10a = 1/dc.T1(3.0, 'blood'), ... R10v = 1/dc.T1(3.0, 'blood'), ... )
Train the model on the ROI data:
>>> model.train(time, roi)
Plot the reconstructed signals (left) and concentrations (right) and compare the concentrations against the noise-free ground truth. Since the data are analysed with an exact model, and there are no other data errors present, this should fior the data exactly.
>>> model.plot(time, roi, ref=gt)
(
Source code,png,hires.png,pdf)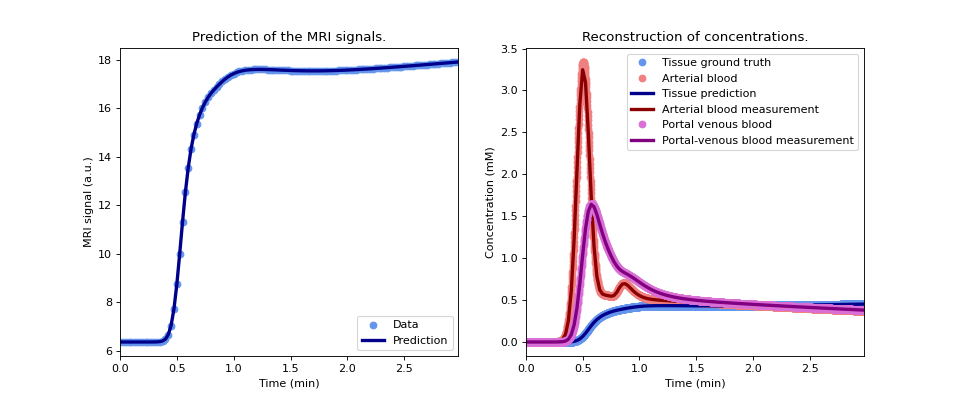
Notes
Table Liver parameters lists the parameters that are relevant in each regime. Table Liver parameter defaults list all possible parameters and their default settings.
Liver parameters# Parameters
When to use
Further detail
n0
Always
For estimating baseline signal
field_strength, agent, R10
Always
R10a, B1corr_a
When aif is provided
R10v, B1corr_v
When vif is provided
S0, FA, TR, TS, B1corr
Always
TP, TC
If sequence is ‘SR’
ve, Fp, fa, Ta, Tg, khe, khe_i, kh_f, Th, Th_i, Th_f.
Depends on kinetics and stationary
Liver parameter defaults# Parameter
Type
Value
Bounds
Free/Fixed
field_strength
Injection
3
[0, inf]
Fixed
agent
Injection
‘gadoxetate’
None
Fixed
R10
Relaxation
0.7
[0, inf]
Fixed
R10a
Relaxation
0.7
[0, inf]
Fixed
R10v
Relaxation
0.7
[0, inf]
Fixed
B1corr
Sequence
1
[0, inf]
Fixed
B1corr_a
Sequence
1
[0, inf]
Fixed
B1corr_v
Sequence
1
[0, inf]
Fixed
FA
Sequence
15
[0, inf]
Fixed
S0
Sequence
1
[0, inf]
Fixed
TC
Sequence
0.1
[0, inf]
Fixed
TP
Sequence
0
[0, inf]
Fixed
TR
Sequence
0.005
[0, inf]
Fixed
TS
Sequence
0
[0, inf]
Fixed
H
Kinetic
0.45
[0, 1]
Fixed
Te
Kinetic
30
[0.1, 60]
Free
De
Kinetic
0.85
[0, 1]
Free
ve
Kinetic
0.3
[0.01, 0.6]
Free
Ta
Kinetic
2
[0, inf]
Free
Tg
Kinetic
10
[0, inf]
Free
Fp
Kinetic
0.008
[0, inf]
Free
fa
Kinetic
0.2
[0, inf]
Free
khe
Kinetic
0.003
[0, 0.1]
Free
khe_i
Kinetic
0.003
[0, 0.1]
Free
khe_f
Kinetic
0.003
[0, 0.1]
Free
Th
Kinetic
1800
[600, 36000]
Free
Th_i
Kinetic
1800
[600, 36000]
Free
Th_f
Kinetic
1800
[600, 36000]
Free
vol
Kinetic
1000
[0, 10000]
Free
Methods
conc([sum])Tissue concentrations
cost(xdata, ydata[, metric])Return the goodness-of-fit
Return model parameters with their descriptions
load([file, path, filename])Load the saved state of the model
params(*args[, round_to])Return the parameter values
plot(time, signal[, ref, xlim, fname, show])Plot the model fit against data
predict(time)Predict the data at specific time points
print_params([round_to])Print the model parameters and their uncertainties
relax()Tissue relaxation rate
save([file, path, filename])Save the current state of the model
set_free([pop])Set the free model parameters.
signal()Pseudocontinuous signal
time()Array of time points.
train(time, signal, **kwargs)Train the free parameters
- conc(sum=True)[source]#
Tissue concentrations
- Parameters:
sum (bool, optional) – If True, returns the total concentrations. If False, returns concentration in both compartments separately. Defaults to True.
- Returns:
Concentration in M
- Return type:
- cost(xdata, ydata, metric='NRMS') float#
Return the goodness-of-fit
- Parameters:
xdata (array-like) – Array with x-data (time points).
ydata (array-like) – Array with y-data (signal values)
metric (str, optional) – Which metric to use (see notes for possible values). Defaults to ‘NRMS’.
- Returns:
goodness of fit.
- Return type:
Notes
Available options are:
‘RMS’: Root-mean-square.
‘NRMS’: Normalized root-mean-square.
‘AIC’: Akaike information criterion.
‘cAIC’: Corrected Akaike information criterion for small models.
‘BIC’: Baysian information criterion.
- export_params() list#
Return model parameters with their descriptions
- Returns:
Dictionary with one item for each model parameter. The key is the parameter symbol (short name), and the value is a 4-element list with [parameter name, value, unit, sdev].
- Return type:
- load(file=None, path=None, filename='Model')#
Load the saved state of the model
- Parameters:
file (str, optional) – complete path of the file. If this is not provided, a file is constructure from path and filename variables. Defaults to None.
path (str, optional) – path to store the state if file is not provided. Thos variable is ignored if file is provided. Defaults to current working directory.
filename (str, optional) – filename to store the state if file is not provided. If no extension is included, the extension ‘.pkl’ is automatically added. This variable is ignored if file is provided. Defaults to ‘Model’.
- Returns:
class instance
- Return type:
- params(*args, round_to=None)#
Return the parameter values
- plot(time: ndarray, signal: ndarray, ref=None, xlim=None, fname=None, show=True)[source]#
Plot the model fit against data
- Parameters:
xdata (array-like) – Array with x-data (time points)
ydata (array-like) – Array with y-data (signal data)
xlim (array_like, optional) – 2-element array with lower and upper boundaries of the x-axis. Defaults to None.
ref (tuple, optional) – Tuple of optional test data in the form (x,y), where x is an array with x-values and y is an array with y-values. Defaults to None.
fname (path, optional) – Filepath to save the image. If no value is provided, the image is not saved. Defaults to None.
show (bool, optional) – If True, the plot is shown. Defaults to True.
- predict(time: ndarray)[source]#
Predict the data at specific time points
- Parameters:
time (array-like) – Array of time points.
- Returns:
Array of predicted data for each element of time.
- Return type:
np.ndarray
- print_params(round_to=None)#
Print the model parameters and their uncertainties
- Parameters:
round_to (int, optional) – Round to how many digits. If this is not provided, the values are not rounded. Defaults to None.
- save(file=None, path=None, filename='Model')#
Save the current state of the model
- Parameters:
file (str, optional) – complete path of the file. If this is not provided, a file is constructure from path and filename variables. Defaults to None.
path (str, optional) – path to store the state if file is not provided. Thos variable is ignored if file is provided. Defaults to current working directory.
filename (str, optional) – filename to store the state if file is not provided. If no extension is included, the extension ‘.pkl’ is automatically added. This variable is ignored if file is provided. Defaults to ‘Model’.
- Returns:
class instance
- Return type:
- set_free(pop=None, **kwargs)#
Set the free model parameters.
- Parameters:
pop (str or list) – a single variable or a list of variables to remove from the list of free parameters.
- Raises:
ValueError – if the pop argument contains a parameter that is not in the list of free parameters.
ValueError – If the parameter is not a model parameter, or bounds are not properly formatted.
- signal() ndarray[source]#
Pseudocontinuous signal
- Returns:
the signal as a 1D array.
- Return type:
np.ndarray
- train(time, signal, **kwargs)[source]#
Train the free parameters
- Parameters:
time (array-like) – Array with time points
signal (array-like) – Array with signal values
kwargs – any keyword parameters accepted by
scipy.optimize.curve_fit, except for bounds.
- Returns:
A reference to the model instance.
- Return type:
