dcmri.KidneyCortMed#
- class dcmri.KidneyCortMed(sequence='SR', **params)[source]#
General model for renal cortico-medullary data.
Kinetic parameters
Hct (float, optional): Hematocrit.
Fp Plasma flow in mL/sec/mL.
Eg Glomerular extraction fraction.
fc Cortical flow fraction.
Tg Glomerular mean transit time in sec.
Tv Peritubular & venous mean transit time in sec.
Tpt Proximal tubuli mean transit time in sec.
Tlh Lis of Henle mean transit time in sec.
Tdt Distal tubuli mean transit time in sec.
Tcd Collecting duct mean transit time in sec.
Input function
aif (array-like, default=None): Signal-time curve in a feeding artery. If AIF is set to None, then the parameter ca must be provided (arterial concentrations).
ca (array-like, default=None): Concentration (M) in the arterial input. Must be provided when aif = None, ignored otherwise.
Acquisition parameters
sequence (str, default=’SR’): imaging sequence.
dt (float, optional): Sampling interval of the AIF in sec.
agent (str, optional): Contrast agent generic name.
field_strength (float, optional): Magnetic field strength in T.
TR (float, optional): Repetition time, or time between excitation pulses, in sec.
FA (float, optional): Nominal flip angle in degrees.
Tsat (float, optional): Time before start of readout (sec).
TC (float, optional): Time to the center of the readout pulse
n0 (float, optional): Baseline length in number of acquisitions.
Signal parameters
R10c (float, optional): Precontrast cortex relaxation rate in 1/sec.
R10m (float, optional): Precontrast medulla relaxation rate in 1/sec.
R10a (float, optional): Precontrast arterial relaxation rate in 1/sec.
S0c (float, optional): Signal scaling factor in the cortex (a.u.).
S0m (float, optional): Signal scaling factor in the medulla (a.u.).
Prediction and training parameters
free (array-like): list of free parameters. The default depends on the kinetics parameter.
free (array-like): 2-element list with lower and upper free of the free parameters. The default depends on the kinetics parameter.
Other parameters
vol (float, optional): Liver volume in mL.
Example
Derive model parameters from simulated data:
>>> import dcmri as dc
Use
fake_kidneyto generate synthetic test data:>>> time, aif, roi, gt = dc.fake_kidney(CNR=100)
Build a tissue model and set the constants to match the experimental conditions of the synthetic test data:
>>> model = dc.KidneyCortMed( ... aif = aif, ... dt = time[1], ... agent = 'gadoterate', ... TR = 0.005, ... FA = 15, ... TC = 0.2, ... n0 = 10, ... )
Train the model on the ROI data and predict signals and concentrations:
>>> model.train(time, roi)
Plot the reconstructed signals (left) and concentrations (right) and compare the concentrations against the noise-free ground truth:
>>> model.plot(time, roi, ref=gt)
(
Source code,png,hires.png,pdf)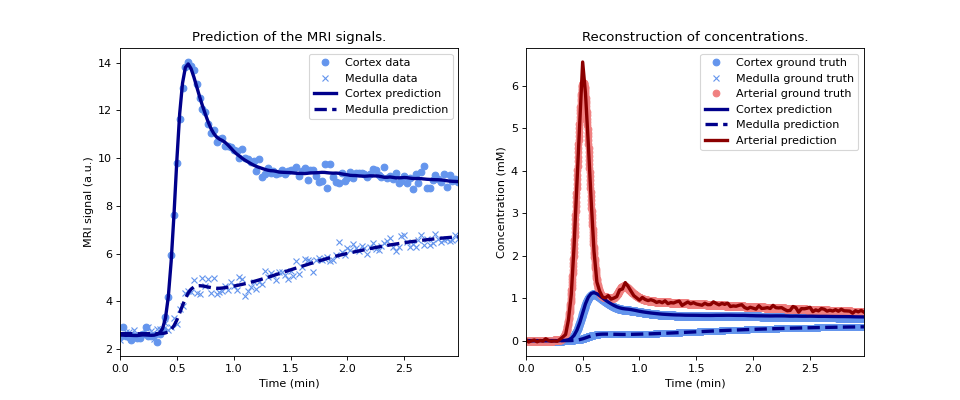
Attributes
lower- and upper free for all free parameters.
Methods
conc([sum])Cortical and medullary concentration
cost(xdata, ydata[, metric])Return the goodness-of-fit
Return model parameters with their descriptions
load([file, path, filename])Load the saved state of the model
params(*args[, round_to])Return the parameter values
plot(xdata, ydata[, ref, xlim, fname, show])Plot the model fit against data
predict(xdata)Predict the data at given xdata
print_params([round_to])Print the model parameters and their uncertainties
save([file, path, filename])Save the current state of the model
set_free([pop])Set the free model parameters.
time()Array of time points
train(xdata, ydata, **kwargs)Train the free parameters
- conc(sum=True)[source]#
Cortical and medullary concentration
- Returns:
Concentration in cortex, concentration in medulla, in M
- Return type:
- cost(xdata: ndarray, ydata: tuple[ndarray, ndarray], metric='NRMS') float[source]#
Return the goodness-of-fit
- Parameters:
xdata (array-like) – Array with x-data (time points).
ydata (tuple) – Tuple of two 1D arrays (signal_cortex, signal_mdeulla) with signals in cortex and medulla, respectively.
metric (str, optional) – Which metric to use - options are: RMS (Root-mean-square); NRMS (Normalized root-mean-square); AIC (Akaike information criterion); cAIC (Corrected Akaike information criterion for small models); BIC (Baysian information criterion). Defaults to ‘NRMS’.
- Returns:
goodness of fit.
- Return type:
- export_params()[source]#
Return model parameters with their descriptions
- Returns:
Dictionary with one item for each model parameter. The key is the parameter symbol (short name), and the value is a 4-element list with [parameter name, value, unit, sdev].
- Return type:
- free = {}#
lower- and upper free for all free parameters.
- load(file=None, path=None, filename='Model')#
Load the saved state of the model
- Parameters:
file (str, optional) – complete path of the file. If this is not provided, a file is constructure from path and filename variables. Defaults to None.
path (str, optional) – path to store the state if file is not provided. Thos variable is ignored if file is provided. Defaults to current working directory.
filename (str, optional) – filename to store the state if file is not provided. If no extension is included, the extension ‘.pkl’ is automatically added. This variable is ignored if file is provided. Defaults to ‘Model’.
- Returns:
class instance
- Return type:
- params(*args, round_to=None)#
Return the parameter values
- plot(xdata: ndarray, ydata: tuple[ndarray, ndarray], ref=None, xlim=None, fname=None, show=True)[source]#
Plot the model fit against data
- Parameters:
xdata (array-like) – Array with x-data (time points)
ydata (tuple of arrays) – Tuple of two 1D arrays (signal_cortex, signal_mdeulla) with signals in cortex and medulla, respectively.
xlim (array_like, optional) – 2-element array with lower and upper boundaries of the x-axis. Defaults to None.
ref (tuple, optional) – Tuple of optional test data in the form (x,y), where x is an array with x-values and y is an array with y-values. Defaults to None.
fname (path, optional) – Filepath to save the image. If no value is provided, the image is not saved. Defaults to None.
show (bool, optional) – If True, the plot is shown. Defaults to True.
- predict(xdata: ndarray) tuple[source]#
Predict the data at given xdata
- Parameters:
xdata (array-like) – Either an array with x-values (time points) or a tuple with multiple such arrays
- Returns:
tuple of two 1D arrays with signal in corex and medulla, respectively.
- Return type:
tuple[np.ndarray,np.ndarray]
- print_params(round_to=None)#
Print the model parameters and their uncertainties
- Parameters:
round_to (int, optional) – Round to how many digits. If this is not provided, the values are not rounded. Defaults to None.
- save(file=None, path=None, filename='Model')#
Save the current state of the model
- Parameters:
file (str, optional) – complete path of the file. If this is not provided, a file is constructure from path and filename variables. Defaults to None.
path (str, optional) – path to store the state if file is not provided. Thos variable is ignored if file is provided. Defaults to current working directory.
filename (str, optional) – filename to store the state if file is not provided. If no extension is included, the extension ‘.pkl’ is automatically added. This variable is ignored if file is provided. Defaults to ‘Model’.
- Returns:
class instance
- Return type:
- set_free(pop=None, **kwargs)#
Set the free model parameters.
- Parameters:
pop (str or list) – a single variable or a list of variables to remove from the list of free parameters.
- Raises:
ValueError – if the pop argument contains a parameter that is not in the list of free parameters.
ValueError – If the parameter is not a model parameter, or bounds are not properly formatted.
- train(xdata: ndarray, ydata: tuple[ndarray, ndarray], **kwargs)[source]#
Train the free parameters
- Parameters:
xdata (array-like) – Array with x-data (time points)
ydata (tuple) – Tuple of two 1D arrays (signal_cortex, signal_mdeulla) with signals in cortex and medulla, respectively.
kwargs – any keyword parameters accepted by
scipy.optimize.curve_fit.
- Returns:
A reference to the model instance.
- Return type:
Model
