dcmri.conc_kidney#
- dcmri.conc_kidney(ca: ndarray, *params, t=None, dt=1.0, sum=True, kinetics='2CF', **kwargs) ndarray[source]#
Concentration in kidney tissues.
- Parameters:
ca (array-like) – concentration in the arterial input.
params (tuple) – free model parameters.
t (array_like, optional) – the time points in sec of the input function ca. If t is not provided, the time points are assumed to be uniformly spaced with spacing dt. Defaults to None.
dt (float, optional) – spacing in seconds between time points for uniformly spaced time points. This parameter is ignored if t is explicity provided. Defaults to 1.0.
kinetics (str, optional) – Kinetics of the tissue, either ‘2CF’, ‘FN’ - see below for detail. Defaults to ‘2CF’.
sum (bool, optional) – For two-compartment tissues, set to True to return the total tissue concentration. Defaults to True.
kwargs (dict, optional) – any optional keyword parameters required by the kinetic model - see below for detail.
- Returns:
If sum=True, this is a 1D array with the total concentration at each time point. If sum=False this is the concentration in each compartment, and at each time point, as a 2D array with dimensions (2,k), where k is the number of time points in ca. The concentration is returned in units of M.
- Return type:
Notes
Currently implemented kinetic models are:
‘2CF’: two-compartment filtration model. params = (Fp, Tp, Ft, Tt,)
‘FN’: free nephron model. params = (Fp, Tp, Ft, h, ).
The model parameters are:
Fp (float, mL/sec/mL): Plasma flow.
Tp (float, sec): plasma mean transit time.
Ft (float, mL/sec/mL): tubular flow.
Tt (float, sec): tubular mean transit time.
hh (array-like, 1/sec): frequences of transit time histogram. The boundaries of the transit time bins can be provided as an array in a keyword parameter TT, which has to have one more element than h. If TT is not provided, the transit time bins are equally space in the range [0, tmax], where tmax is the largest acquisition time.
Example
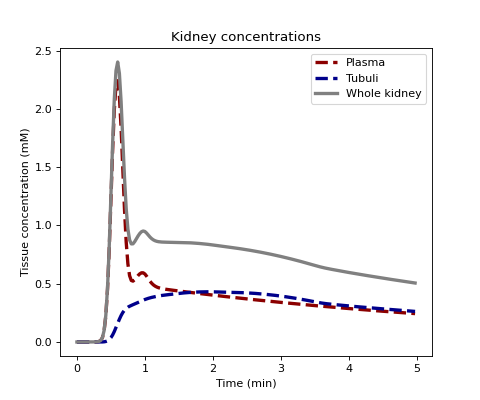
Plot concentration in cortex and medulla for typical values:
>>> import matplotlib.pyplot as plt >>> import numpy as np >>> import dcmri as dc
Generate a population-average input function:
>>> t = np.arange(0, 300, 1.5) >>> ca = dc.aif_parker(t, BAT=20)
Define some parameters and generate plasma and tubular tissue concentrations with a 2-compartment filtration model:
>>> Fp, Tp, Ft, Tt = 0.05, 10, 0.01, 120 >>> C = dc.conc_kidney(ca, Fp, Tp, Ft, Tt, t=t, sum=False, kinetics='2CF')
Plot all concentrations:
>>> fig, ax = plt.subplots(1,1,figsize=(6,5)) >>> ax.set_title('Kidney concentrations') >>> ax.plot(t/60, 1000*C[0,:], linestyle='--', linewidth=3.0, color='darkred', label='Plasma') >>> ax.plot(t/60, 1000*C[1,:], linestyle='--', linewidth=3.0, color='darkblue', label='Tubuli') >>> ax.plot(t/60, 1000*(C[0,:]+C[1,:]), linestyle='-', linewidth=3.0, color='grey', label='Whole kidney') >>> ax.set_xlabel('Time (min)') >>> ax.set_ylabel('Tissue concentration (mM)') >>> ax.legend() >>> plt.show()
(
Source code,png,hires.png,pdf)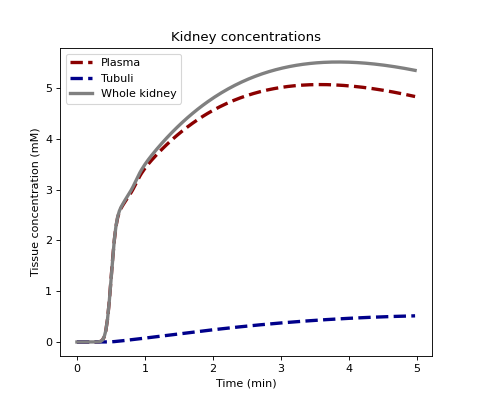
Use generate plasma and tubular tissue concentrations using the free nephron model for comparison. We assume 4 transit time bins with the following boundaries (in units of seconds):
>>> TT = [0, 15, 30, 60, 120]
with longest transit times most likely (note the frequences to not have to add up to 1):
>>> h = [1, 2, 3, 4] >>> C = dc.conc_kidney(ca, Fp, Tp, Ft, h, t=t, sum=False, kinetics='FN', TT=TT)
Plot all concentrations:
>>> fig, ax = plt.subplots(1,1,figsize=(6,5)) >>> ax.set_title('Kidney concentrations') >>> ax.plot(t/60, 1000*C[0,:], linestyle='--', linewidth=3.0, color='darkred', label='Plasma') >>> ax.plot(t/60, 1000*C[1,:], linestyle='--', linewidth=3.0, color='darkblue', label='Tubuli') >>> ax.plot(t/60, 1000*(C[0,:]+C[1,:]), linestyle='-', linewidth=3.0, color='grey', label='Whole kidney') >>> ax.set_xlabel('Time (min)') >>> ax.set_ylabel('Tissue concentration (mM)') >>> ax.legend() >>> plt.show()