dcmri.signal_tissue#
- dcmri.signal_tissue(ca: ndarray, R10: float, r1: float, t=None, dt=1.0, kinetics='2CX', water_exchange='FF', sequence=None, inflow=None, **params) ndarray[source]#
Signal for a 2-site exchange tissue. For more detail see Exchange tissues.
- Parameters:
ca (array-like) – concentration in the blood of the arterial input.
R10 (float) – precontrast relaxation rate. The tissue is assumed to be in fast exchange before injection of contrast agent.
r1 (float) – contrast agent relaxivity.
t (array_like, optional) – the time points in sec of the input function ca. If t is not provided, the time points are assumed to be uniformly spaced with spacing dt. Defaults to None.
dt (float, optional) – spacing in seconds between time points for uniformly spaced time points. This parameter is ignored if t is explicity provided. Defaults to 1.0.
kinetics (str, optional) – Tracer-kinetic model. Possible values are ‘2CX’, ‘2CU’, ‘HF’, ‘HFU’, ‘NX’, ‘FX’, ‘WV’, ‘U’. Defaults to ‘2CX’.
water_exchange (str, optional) – Water exchange regime, Any combination of two of the letters ‘F’, ‘N’, ‘R’ is allowed. Defaults to ‘FF’.
sequence (dict) – the sequence model and its parameters. The dictionary has one required key ‘model’ which specifies the signal model. Currently either ‘SS’ or ‘SR’. The other keys are the values of the signal parameter, which depend on the model. See table Tissue signal parameters for detail.
inflow (dict, optional) – inflow model. If not provided, the in- and outflow of magnetization is ignored. To include inflow effects, inflow must be dictionary with the signal model parameters for the arterial input. For the ‘SS’ signal model, required parameters are ‘R10a’ and ‘B1corr_a’. Defaults to None.
params (dict) – model parameters. See Tissue signal parameters for more detail. Note: the tissue parameters are keyword arguments for convenience, but a value is required.
- Raises:
ValueError – if a required parameter has no value assigned.
NotImplementedError – if a combination of regimes is not yet implemented. Currently the sequence type ‘SR’ only accepts fast water exchange ‘FF’.
- Returns:
tissue signal as a 1D array.
- Return type:
ndarray
Example
We verify that the effect of inflow is negligible in a steady state sequence:
>>> import matplotlib.pyplot as plt >>> import numpy as np >>> import dcmri as dc
Define constants and model parameters:
>>> R10, r1 = 1, 5000 >>> seq = {'model': 'SS', 'S0':1, 'FA':15, 'TR': 0.001, 'B1corr':1} >>> pars = { >>> 'sequence':seq, 'kinetics':'2CX', 'water_exchange':'NN', >>> 'H':0.045, 'vb':0.05, 'vi':0.3, 'Fb':0.01, 'PS':0.005} >>> inflow = {'R10a': 0.7, 'B1corr_a':1}
Generate arterial blood concentrations:
>>> t = np.arange(0, 300, 1.5) >>> ca = dc.aif_parker(t, BAT=20)/(1-0.45)
Calculate the signal with and without inflow:
>>> Sf = dc.signal_tissue(ca, R10, r1, t=t, inflow=inflow, **pars) >>> Sn = dc.signal_tissue(ca, R10, r1, t=t, **pars)
Compare them in a plot:
>>> plt.figure() >>> plt.plot(t/60, Sn, label='Without inflow correction', linewidth=3) >>> plt.plot(t/60, Sf, label='With inflow correction') >>> plt.xlabel('Time (min)') >>> plt.ylabel('Concentration (mM)') >>> plt.legend() >>> plt.show()
(
Source code,png,hires.png,pdf)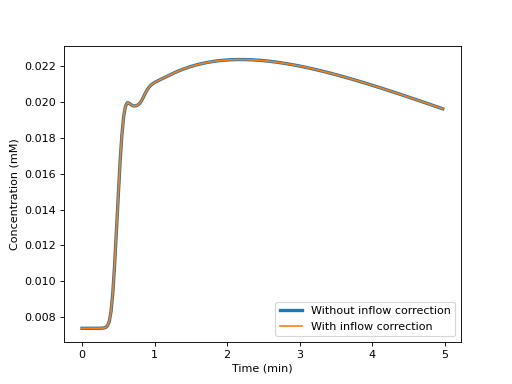
Notes
Tissue signal parameters# Parameters
When to use
Further detail
Fb, PS, Ktrans, vb, H, vi, ve, vc, PSe, PSc.
Depends on kinetics and water_exchange
S0, FA, TR, B1corr
Always
TP, TC
If sequence is ‘SR’
R10a, B1corr_a
If inflow is not None
