dcmri.fetch#
- dcmri.fetch(dataset=None, clear_cache=False, download_all=False) dict[source]#
Fetch a dataset included in dcmri
- Parameters:
dataset (str, optional) – name of the dataset. See below for options.
clear_cache (bool, optional) – When a dataset is fetched, it is downloaded and then stored in a local cache memory for faster access next time it is fetched. Set clear_cache=True to delete all data in the cache memory. Default is False.
download_all (bool, optional) – By default only the dataset that is fetched is downloaded. Set download_all=True to download all datasets at once. This will cost some time but then offers fast and offline access to all datasets afterwards. This will take up around 300 MB of space on your hard drive. Default is False.
- Returns:
Data as a dictionary.
- Return type:
Notes
The following datasets are currently available:
tristan_rifampicin
Background: data are provided by the liver work package of the TRISTAN project which develops imaging biomarkers for drug safety assessment. The data and analysis was first presented at the ISMRM in 2024 (Min et al 2024, manuscript in press).
The data were acquired in the aorta and liver of 10 healthy volunteers with dynamic gadoxetate-enhanced MRI, before and after administration of a drug (rifampicin) which is known to inhibit liver function. The assessments were done on two separate visits at least 2 weeks apart. On each visit, the volunteer had two scans each with a separate contrast agent injection of a quarter dose each. the scans were separated by a gap of about 1 hour to enable gadoxetate to clear from the liver. This design was deemed necessary for reliable measurement of excretion rate when liver function was inhibited.
The research question was to what extent rifampicin inhibits gadoxetate uptake rate from the extracellular space into the liver hepatocytes (khe, mL/min/100mL) and excretion rate from hepatocytes to bile (kbh, mL/100mL/min). 2 of the volunteers only had the baseline assessment, the other 8 volunteers completed the full study. The results showed consistent and strong inhibition of khe (95%) and kbh (40%) by rifampicin. This implies that rifampicin poses a risk of drug-drug interactions (DDI), meaning it can cause another drug to circulate in the body for far longer than expected, potentially causing harm or raising a need for dose adjustment.
Data format: The fetch function returns a list of dictionaries, one per subject visit. Each dictionary contains the following items:
time1aorta: array of signals in arbitrary units, for the aorta in the first scan.
time2aorta: array of signals in arbitrary units, for the aorta in the second scan.
time1liver: array of signals in arbitrary units, for the liver in the first scan.
time2liver: array of signals in arbitrary units, for the liver in the second scan.
signal1aorta: array of signals in arbitrary units, for the aorta in the first scan.
signal2aorta: array of signals in arbitrary units, for the aorta in the second scan.
signal1liver: array of signals in arbitrary units, for the liver in the first scan.
signal2liver: array of signals in arbitrary units, for the liver in the second scan.
weight: subject weight in kg.
agent: contrast agent generic name (str).
dose: 2-element list with contrast agent doses of first scan and second scan in mL/kg.
rate: contrast agent injection rate in mL/sec.
FA: Flip angle in degrees
TR: repretition time in sec
t0: baseline length in subject
subject: Volunteer number.
visit: either ‘baseline’ or ‘rifampicin’.
field_strength: B0-field of scanner.
R10b: precontrast R1 of blood (1st scan).
R10l: precontrast R1 of liver (1st scan).
R102b: precontrast R1 of blood (2nd scan).
R102l: precontrast R1 of liver (2nd scan).
Hct: hematocrit.
vol: liver volume in mL.
Please reference the following abstract when using these data:
Thazin Min, Marta Tibiletti, Paul Hockings, Aleksandra Galetin, Ebony Gunwhy, Gerry Kenna, Nicola Melillo, Geoff JM Parker, Gunnar Schuetz, Daniel Scotcher, John Waterton, Ian Rowe, and Steven Sourbron. Measurement of liver function with dynamic gadoxetate- enhanced MRI: a validation study in healthy volunteers. Proc Intl Soc Mag Reson Med, Singapore 2024.
tristan_gothenburg
Background: The data aimed to demonstrates the effect of rifampicin on liver function of patients with impaired function. The data are provided by the liver work package of the TRISTAN project which develops imaging biomarkers for drug safety assessment.
The data were acquired in the aorta and liver in 3 patients with dynamic gadoxetate-enhanced MRI. The study participants take rifampicin as part of their routine clinical workup, with an aim to promote their liver function. For this study, they were taken off rifampicin 3 days before the first scan, and placed back on rifampicin 3 days before the second scan. The aim was to determine the effect if rifampicin in uptake and excretion function of the liver.
The data confirmed that patients had significantly reduced uptake and excretion function in the absence of rifampicin. Rifampicin adminstration promoted their excretory function but had no effect on their uptake function.
Data format: The fetch function returns a list of dictionaries, one per subject visit. Each dictionary contains the following items:
time1aorta: array of signals in arbitrary units, for the aorta in the first scan.
time2aorta: array of signals in arbitrary units, for the aorta in the second scan.
time1liver: array of signals in arbitrary units, for the liver in the first scan.
time2liver: array of signals in arbitrary units, for the liver in the second scan.
signal1aorta: array of signals in arbitrary units, for the aorta in the first scan.
signal2aorta: array of signals in arbitrary units, for the aorta in the second scan.
signal1liver: array of signals in arbitrary units, for the liver in the first scan.
signal2liver: array of signals in arbitrary units, for the liver in the second scan.
weight: subject weight in kg.
agent: contrast agent generic name (str).
dose: 2-element list with contrast agent doses of first scan and second scan in mL/kg.
rate: contrast agent injection rate in mL/sec.
FA: Flip angle in degrees
TR: repretition time in sec
t0: baseline length in subject
subject: Volunteer number.
visit: either ‘control’ or ‘drug’.
field_strength: B0-field of scanner.
R10b: precontrast R1 of blood (1st scan).
R10l: precontrast R1 of liver (1st scan).
R102b: precontrast R1 of blood (2nd scan).
R102l: precontrast R1 of liver (2nd scan).
Hct: hematocrit.
vol: liver volume in mL.
tristan6drugs
Background: data are provided by the liver work package of the TRISTAN project which develops imaging biomarkers for drug safety assessment. The data and analysis were first published in Melillo et al (2023).
The study presented here measured gadoxetate uptake and excretion in healthy rats before and after injection of 6 test drugs (up to 6 rats per drug). Studies were performed in preclinical MRI scanners at 3 different centers and 2 different field strengths.
Results demonstrated that two of the tested drugs (rifampicin and cyclosporine) showed strong inhibition of both uptake and excretion. One drug (ketoconazole) inhibited uptake but not excretion. Three drugs (pioglitazone, bosentan and asunaprevir) inhibited excretion but not uptake.
Data format: The fetch function returns a list of dictionaries, one per scan. Each dictionary contains the following items:
time: array of time points in sec
spleen: array of spleen signals in arbitrary units
liver: array of liver signals in arbitrary units.
FA: Flip angle in degrees
TR: repretition time in sec
n0: number of precontrast acquisitions
study: an integer identifying the substudy the scan was taken in
subject: a study-specific identifier of the subject in the range 1-6.
visit: either 1 (baseline) or 2 (drug or vehicle/saline).
center: center wehere the study was performed, either E, G or D.
field_strength: B0-field of scanner on whuch the study was performed
substance: what was injected, eg. saline, vehicle or drug name.
BAT: Bolus arrival time
duration: duration on the injection in sec.
Please reference the following paper when using these data:
Melillo N, Scotcher D, Kenna JG, Green C, Hines CDG, Laitinen I, Hockings PD, Ogungbenro K, Gunwhy ER, Sourbron S, et al. Use of In Vivo Imaging and Physiologically-Based Kinetic Modelling to Predict Hepatic Transporter Mediated Drug–Drug Interactions in Rats. Pharmaceutics. 2023; 15(3):896. [DOI]
The data were first released as supplementary material in csv format with this paper on Zenodo. Use this DOI to reference the data themselves:
Gunwhy, E. R., & Sourbron, S. (2023). TRISTAN-RAT (v3.0.0). Zenodo
tristan_repro
Background: data are provided by the liver work package of the TRISTAN project which develops imaging biomarkers for drug safety assessment. The data and analysis were first published in Gunwhy et al (2024).
The study presented here aimed to determine the repreducibility and rpeatability of gadoxetate uptake and excretion measurements in healthy rats. Data were acquired in different centers and field strengths to identify contributing factors. Some of the studies involved repeat scans in the same subject. In some studies data on the second day were taken after adminstration of a drug (rifampicin) to test if effect sizes were reproducible.
Data format: The fetch function returns a list of dictionaries, one per scan. The dictionaries in the list contain the following items:
time: array of time points in sec
spleen: array of spleen signals in arbitrary units
liver: array of liver signals in arbitrary units.
FA: Flip angle in degrees
TR: repretition time in sec
n0: number of precontrast acquisitions
study: an integer identifying the substudy the scan was taken in
subject: a study-specific identifier of the subject in the range 1-6.
visit: either 1 (baseline) or 2 (drug or vehicle/saline).
center: center wehere the study was performed, either E, G or D.
field_strength: B0-field of scanner on whuch the study was performed
substance: what was injected, eg. saline, vehicle or drug name.
BAT: Bolus arrival time
duration: duration on the injection in sec.
Please reference the following paper when using these data:
Ebony R. Gunwhy, Catherine D. G. Hines, Claudia Green, Iina Laitinen, Sirisha Tadimalla, Paul D. Hockings, Gunnar Schütz, J. Gerry Kenna, Steven Sourbron, and John C. Waterton. Assessment of hepatic transporter function in rats using dynamic gadoxetate- enhanced MRI: A reproducibility study. In review.
The data were first released as supplementary material in csv format with this paper on Zenodo. Use this to reference the data themselves:
Gunwhy, E. R., Hines, C. D. G., Green, C., Laitinen, I., Tadimalla, S., Hockings, P. D., Schütz, G., Kenna, J. G., Sourbron, S., & Waterton, J. C. (2023). Rat gadoxetate MRI signal dataset for the IMI-WP2-TRISTAN Reproducibility study [Data set]. Zenodo.
tristan_mdosing
Background These data were taken from a preclinical study which aimed to investigate the potential of gadoxetate-enhanced DCE-MRI to study acute inhibition of hepatocyte transporters of drug-induced liver injury (DILI) causing drugs, and to study potential changes in transporter function after chronic dosing.
Data format: The fetch function returns a list of dictionaries, one per scan.The dictionaries in the list contain the following items:
time: array of time points in sec
spleen: array of spleen signals in arbitrary units
liver: array of liver signals in arbitrary units.
FA: Flip angle in degrees
TR: repretition time in sec
n0: number of precontrast acquisitions
study: an integer identifying the substudy the scan was taken in
subject: a study-specific identifier of the subject in the range 1-6.
visit: either 1 (baseline) or 2 (drug or vehicle/saline).
center: center wehere the study was performed, either E, G or D.
field_strength: B0-field of scanner on whuch the study was performed
substance: what was injected, eg. saline, vehicle or drug name.
BAT: Bolus arrival time
duration: duration on the injection in sec.
Please reference the following abstract when using these data:
Mikael Montelius, Steven Sourbron, Nicola Melillo, Daniel Scotcher, Aleksandra Galetin, Gunnar Schuetz, Claudia Green, Edvin Johansson, John C. Waterton, and Paul Hockings. Acute and chronic rifampicin effect on gadoxetate uptake in rats using gadoxetate DCE-MRI. Int Soc Mag Reson Med 2021; 2674.
kruk_sk_gfr
Background: data taken from supplementary material of Basak et al (2018), a study funded by Kidney Research UK. The dataset includes signal-time curves for aorta, left- and right kidney as well as sequence parameters and radio-isotope single kidney GFR values for 114 scans on 100 different subjects. This includes 14 subjects who have had a scan before and after revascularization treatment.
Data format: The fetch function returns a list of dictionaries, one per scan. The dictionaries in the list contain the following items:
subject: unique study ID of the participant
time: array of time points in sec
aorta: signal curve in the aorta (arbitrary units)
visit: for participants that had just a single visit, this has the value ‘single’. For those that had multiple visits, the value is either ‘pre’ (before treatment) or ‘post’ (after treatment).
LK: signal curve in the left kidney (arbitrary units).
RK: signal curve in the right kidney (arbitrary units).
LK vol: volume of the left kidney (mL).
RK vol: volume of the right kidney (mL).
LK iso-SK-GFR: radio-isotope SK-GFR for the left kidney (mL/min).
RK iso-SK-GFR: radio-isotope SK-GFR for the right kidney (mL/min).
LK T1: precontrast T1-value of the left kidney (sec).
RK T1: precontrast T1-value of the right kidney (sec).
TR: repetition time or time between rf-pulses (sec)
FA: flip angle (degrees)
n0: number of precontrast acquisitions.
field_strength: Magnetic field strength of the scanner (T)
agent: Contrast agent generic name
dose: contrast agent dose injected (mmol/kg)
rate: rate of contrast agent injection (mL/sec)
weight: participant weight (kg)
Note: if data are missing for a particular scan, they will not be in the dictionary for that scan. For instance, if a participant does not have a right kidney, the items starting with RK are not present.
Please reference the following paper when using these data:
Basak S, Buckley DL, Chrysochou C, Banerji A, Vassallo D, Odudu A, Kalra PA, Sourbron SP. Analytical validation of single-kidney glomerular filtration rate and split renal function as measured with magnetic resonance renography. Magn Reson Imaging. 2019 Jun;59:53-60. doi: 10.1016/j.mri.2019.03.005. [URL].
Example:
Use the AortaLiver model to fit one of the tristan_rifampicin datasets:
>>> import dcmri as dc
Get the data for the baseline visit of the first subject in the study:
>>> data = dc.fetch('tristan_rifampicin') >>> data = data[0]
Initialize the AortaLiver model with the available data:
>>> model = dc.AortaLiver( >>> # >>> # Injection parameters >>> # >>> weight = data['weight'], >>> agent = data['agent'], >>> dose = data['dose'][0], >>> rate = data['rate'], >>> # >>> # Acquisition parameters >>> # >>> field_strength = data['field_strength'], >>> t0 = data['t0'], >>> TR = data['TR'], >>> FA = data['FA'], >>> # >>> # Signal parameters >>> # >>> R10a = data['R10b'], >>> R10l = data['R10l'], >>> # >>> # Tissue parameters >>> # >>> H = data['Hct'], >>> vol = data['vol'], >>> )
We are only fitting here the first scan data, so the xdata are the aorta- and liver time points of the first scan, and the ydata are the signals at these time points:
>>> xdata = (data['time1aorta'], data['time1liver']) >>> ydata = (data['signal1aorta'], data['signal1liver'])
Train the model using these data and plot the results to check that the model has fitted the data:
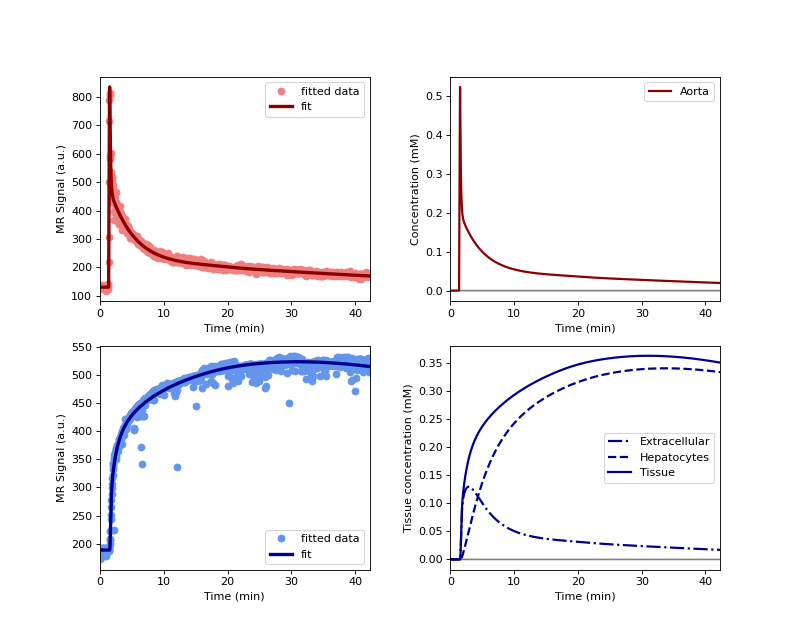
>>> model.train(xdata, ydata, xtol=1e-3) >>> model.plot(xdata, ydata)
(
Source code,png,hires.png,pdf)