dcmri.TissueLS#
- class dcmri.TissueLS(sequence='SS', **kwargs)[source]#
Array of linear and stationary tissues with a single inlet.
These are generic model-free tissue types. Their response to an indicator injection is proportional to the dose (linear) and independent of the time of injection (stationary).
- Parameters:
shape (array-like, required) – shape of the tissue array (spatial dimensions only). Any number of dimensions is allowed.
aif (array-like, required) – Signal-time curve in the blood of the feeding artery.
dt (float, optional) – Time interval between values of the arterial input function. Defaults to 1.0.
sequence (str, optional) – imaging sequence. Possible values are ‘SS’, ‘SR’ and ‘lin’ (linear). Defaults to ‘SS’.
params (dict, optional) – values for the parameters of the tissue, specified as keyword parameters. Defaults are used for any that are not provided.
See also
Example
Fit a linear and stationary model to the synthetic test data:
>>> import numpy as np >>> import dcmri as dc
Generate synthetic test data:
>>> time, aif, roi, gt = dc.fake_tissue()
The correct ground truth for ve in model-free analysis is the extracellular part of the distribution space:
>>> gt['ve'] = gt['vp'] + gt['vi'] if gt['PS'] > 0 else gt['vp']
Build a tissue and set the constants to match the experimental conditions of the synthetic test data.
>>> tissue = dc.TissueLS( ... dt = time[1], ... sequence = 'SS', ... r1 = dc.relaxivity(3, 'blood','gadodiamide'), ... TR = 0.005, ... FA = 15, ... R10a = 1/dc.T1(3.0,'blood'), ... R10 = 1/dc.T1(3.0,'muscle'), ... )
Train the tissue on the data. Since have noise-free synthetic data we use a lower tolerance than the default, which is optimized for noisy data:
>>> tissue.train(roi, aif, n0=10, tol=0.01)
Plot the reconstructed signals along with the concentrations and the impulse response function.
>>> tissue.plot(roi)
(
Source code,png,hires.png,pdf)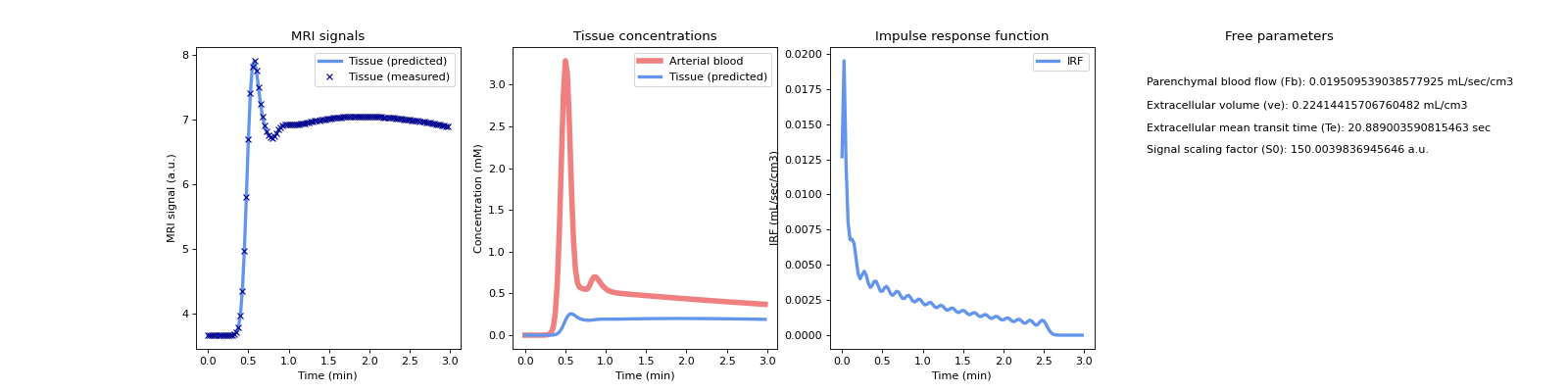
Methods
params(*args)Export the tissue parameters
plot([signal, round_to, fname, show])Plot the model fit against data
predict()Predict the signal at specific time points
Predict the signal at specific time points
Return the tissue concentration
print_params([round_to])Print the model parameters
train(signal, signal_aif[, n0, tol, init_s0])Train the free parameters
- plot(signal=None, round_to=None, fname=None, show=True)[source]#
Plot the model fit against data
- Parameters:
signal (array-like, optional) – Array with measured signals.
round_to (int, optional) – Rounding for the model parameters.
fname (path, optional) – Filepath to save the image. If no value is provided, the image is not saved. Defaults to None.
show (bool, optional) – If True, the plot is shown. Defaults to True.
- predict()[source]#
Predict the signal at specific time points
- Returns:
Array of predicted signals for each time point.
- Return type:
np.ndarray
- predict_aif()[source]#
Predict the signal at specific time points
- Returns:
Array of predicted signals for each time point.
- Return type:
np.ndarray
- predict_conc()[source]#
Return the tissue concentration
- Returns:
Concentration in M
- Return type:
np.ndarray
