dcmri.Mz_spgr#
- dcmri.Mz_spgr(R1, T, TR, FA, TP=0, v=1, Fw=0, j=None, n0=0, me=1)[source]#
Longitudinal tissue magnetization of a spoiled gradient-echo sequence.
See section Longitudinal relaxation for more detail.
- Parameters:
R1 (array-like) – Longitudinal relaxation rates in 1/sec. For a tissue with n compartments, the first dimension of R1 must be n. For a single compartment, R1 can be scalar or a 1D time-array.
T (float) – time since the first rf-pulse.
TR (float) – repetition time between rf-pulses.
FA (float) – flip angle of the rf-pulse.
TP (float, optional) – preparation delay. Default is 0.
v (array-like, optional) – volume fractions of the compartments. For a one-compartment tissue this is a scalar - otherwise it is an array with one value for each compartment. Defaults to 1.
Fw (array-like, optional) – Water flow between the compartments and to the environment, in units of mL/sec/cm3. Generally Fw must be a nxn array, where n is the number of compartments, and the off-diagonal elements Fw[j,i] are the permeability for water moving from compartment i into j. The diagonal elements Fw[i,i] quantify the flow of water from compartment i to outside. For a closed system with equal permeabilities between all compartments, a scalar value for Fw can be provided. Defaults to 0.
j (array-like, optional) – normalized tissue magnetization flux. j has to have the same shape as R1. Defaults to None.
n0 (array-like, optional) – initial relative magnetization at T=0. If this is a scalar, all compartments are assumed to have the same initial magnetization. Defaults to 0.
me (array-like, optional) – equilibrium magnetization of the tissue compartments. If a scalar value is provided, all compartments are assumed to have the same equilibrium magnetization. Defaults to 1.
- Returns:
Tissue magnetization in the compartments after a time T.
- Return type:
np.ndarray
Example
Compare SPGR tissue magnetization of a two-compartment tissue after inversion against free recovery and steady-state:
Import packages:
>>> import numpy as np >>> import matplotlib.pyplot as plt >>> import dcmri as dc
Define constants:
>>> FA, TR, TP = 12, 0.005, 0 >>> TI = np.linspace(0,3,100) >>> R1 = [1, 0.5] >>> v = [0.3, 0.7] >>> f, PS = 0.5, 0.1 >>> Fw = [[f, PS], [PS, 0]]
Compute magnetization:
>>> Mspgr = dc.Mz_spgr(R1, TI, TR, FA, TP, v, Fw, j=[f, 0], n0=-1) >>> Mfree = dc.Mz_free(R1, TI, v, Fw, j=[f, 0], n0=-1) >>> Mss = dc.Mz_ss(R1, TR, FA, v, Fw, j=[f, 0])
Plot the results for the peripheral compartment:
>>> c = 1 >>> plt.title('Peripheral compartment') >>> plt.plot(TI, Mfree[c,:], label='Free', linewidth=3) >>> plt.plot(TI, v[c]+0*TI, label='Equilibrium', linewidth=3) >>> plt.plot(TI, Mspgr[c,:], label='SPGR', linewidth=3) >>> plt.plot(TI, Mss[c]+0*TI, label='Steady-state', linewidth=3) >>> plt.xlabel('Time since start of pulse sequence (sec)') >>> plt.ylabel('Tissue magnetization (A/cm/cm3)') >>> plt.legend() >>> plt.show()
(
Source code,png,hires.png,pdf)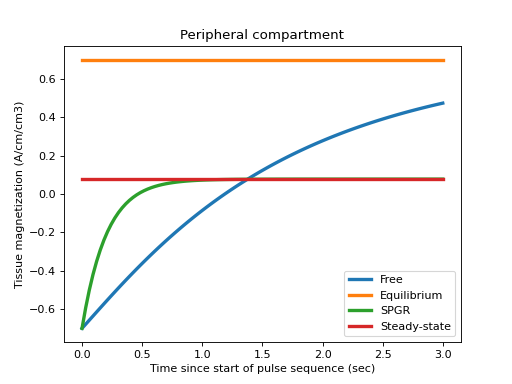
This verifies that the free recovery magnetization goes to equilibrium, and that the SPGR magnetization relaxes to the steady state at a shorter time than the free recovery.
